This tutorial shows how FHIR RDF medical records using SNOMED-CT codes can be processed by a reasoner to identify diagnoses that were not directly coded. Two examples are demonstrated: a FHIR DiagnosticReport for malignant neoplasm is inferred to be an instance of CancerDiagnosis; and a report of a thyroid tumor is inferred to be a thyroid disease diagnosis.
This approach is useful in primary and secondary care institutions to count or identify patients that belong to a particular group of diagnoses. Instead of explicitly querying for every possible code that would indicate the target diagnosis – such as cancer or thyroid disease – the reasoner uses SNOMED-CT’s ontology to infer that diagnosis based on subclass relationships.
This tutorial is based on a Yosemite Project webinar and a paper from the 2017 International SWAT4HCLS Conference by Harold R. Solbrig of Mayo Clinic.
Watch the video of this tutorial (90 seconds)
Target audience
Anyone interested in using FHIR/RDF to perform inference using the SNOMED-CT ontology.
Prerequisites
- Familiarity with semantic web technologies and standards, including OWL, RDF, and the Protege ontology editor
- Familiarity with health informatics standards, including SNOMED-CT and HL7 FHIR
-
A current version of Protege. This tutorial was tested on 5.5.0beta9.
For convenience we will use Protege with the FaCT++ reasoner to perform the inference that will identify a FHIR patient record as a cancer diagnosis. However, if you wanted to perform this inference in a production system you probably would not do it in Protege.
Steps
-
Clone a copy of this repository, which contains the files for this tutorial, and change to that directory.
git clone https://github.com/yosemiteproject/Tutorial-FHIR-RDF-as-a-Bridge.git cd Tutorial-FHIR-RDF-as-a-Bridge -
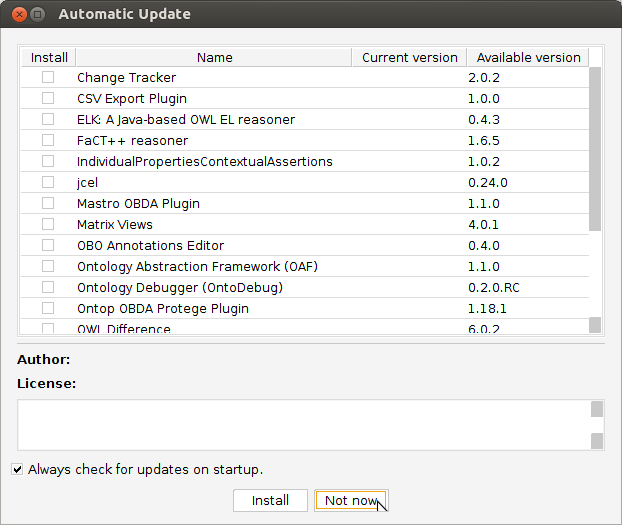
Start Protege. If you get an “Automatic Update” dialog, you may dismiss it by clicking “Not now”.
-
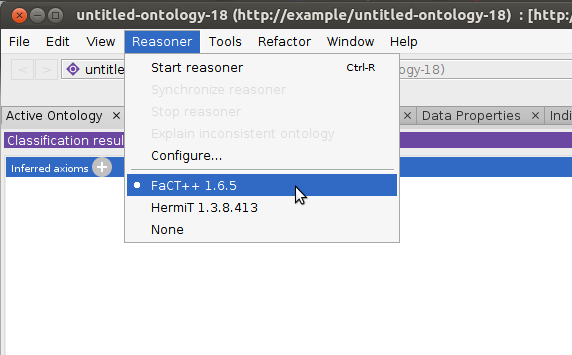
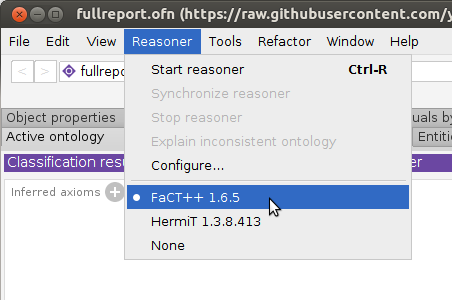
Verify that the FaCT++ reasoner is installed: click the Reasoner menu to see if FaCT++ is listed. If not, install it directly from Protege: File–>Check for plugins…, check “FaCT++ reasoner”, Install, then exit and restart Protege.
NOTE: A user has reported: “The default preferences in Protege don’t display inferred instances, so you might want to add a note to change that after installing or users will never see classified instances.” 25-Nov-2019
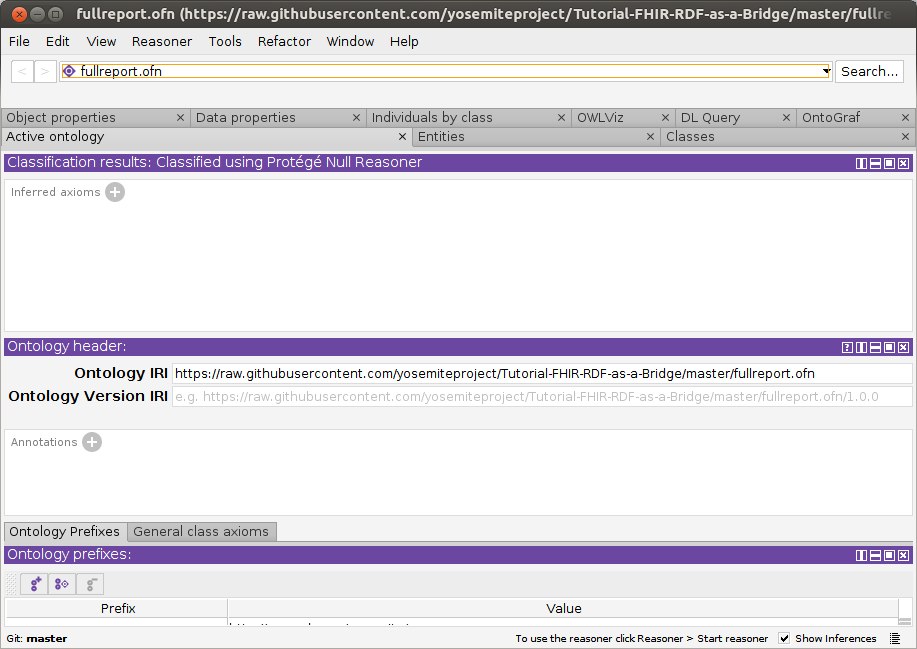
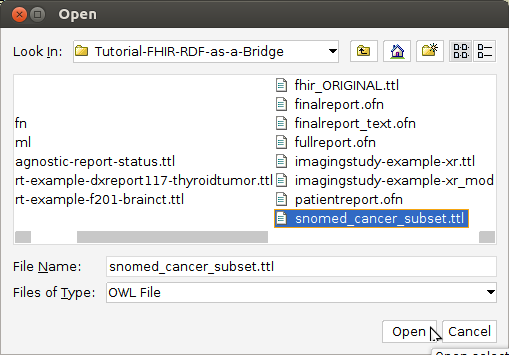
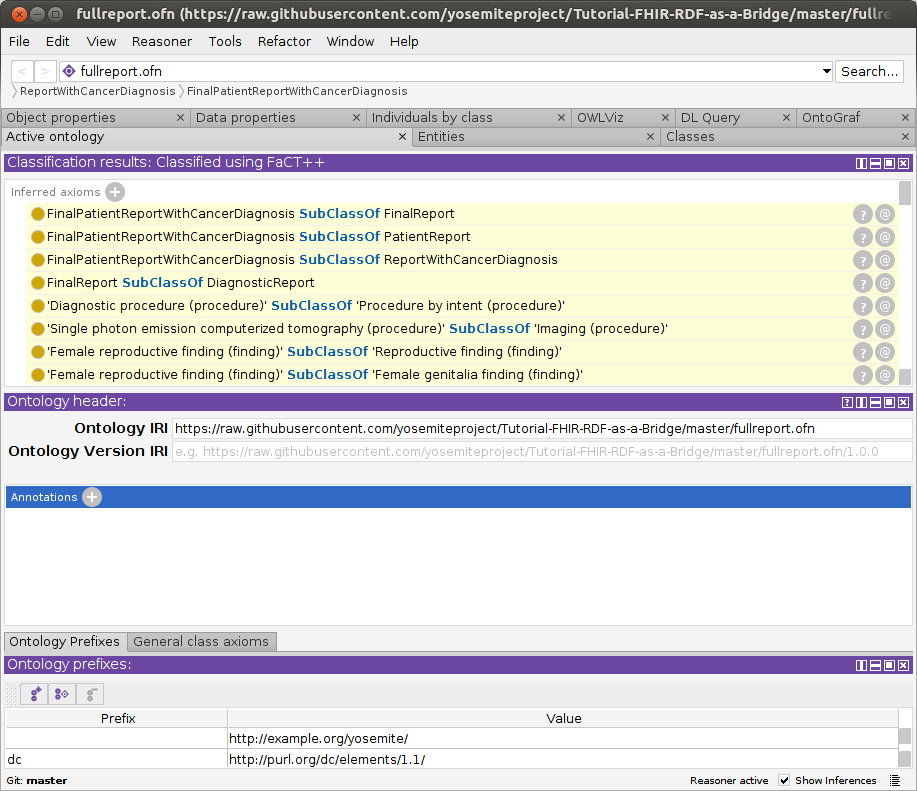
- Open fullreport.ofn: File–>Open.
This OWL file references a sample FHIR/RDF patient data record (f201.ttl)
that we will identify as a cancer diagnosis, using the FaCT++ reasoner. It also references the various FHIR and SNOMED-CT ontology pieces that enable
the reasoner to reach this conclusion. If needed, resolve missing imports using these local files:
snomed_cancer_subset.ttl fhir.ttl w5.ttl diagnosticreport-example-f201-brainct.ttl patientreport.ofn cancerreport.ofn finalreport.ofn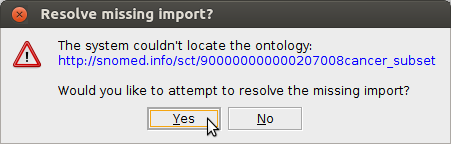
-
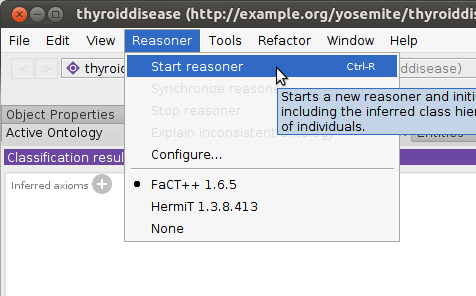
Select the FaCT++ reasoner under the
Reasonermenu.
-
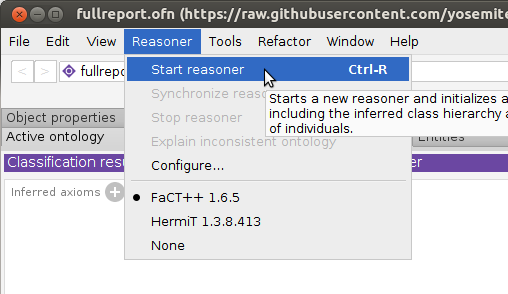
Select
Start Reasonerunder theReasonermenu. It took ~30 seconds to run on a 3.4GHz laptop.
-
After the reasoner has finished, navigate to
FinalPatientReportWithCancerDiagnosisin theClasses–>Class hierarchytab and observe thatf201(the id of the DiagnosticReport) has been recognized as an instance. Success! This means that the reasoner has concluded that this patient record (f201) has a cancer diagnosis.
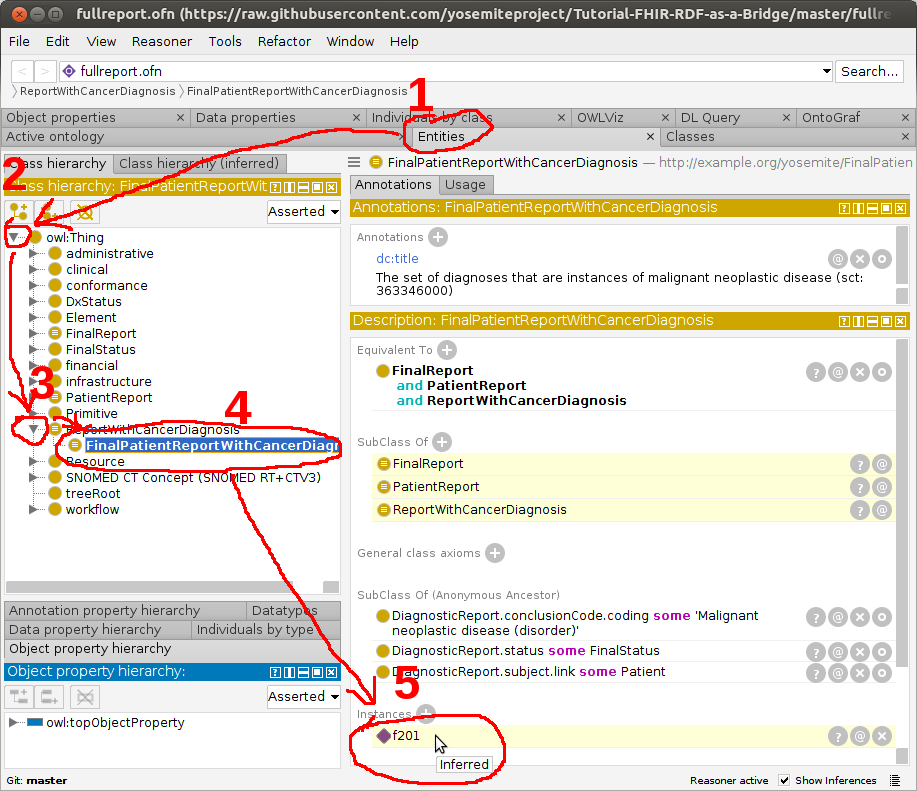
Next, we will test a different patient record for a thyroid disease diagnosis.
-
Open thyroidreport.ofn, answering “no” to the current window prompt. Again, this file imports the ontologies that we need, imports the patient record that will be tested (diagnosticreport-example-dxreport117-thyroidtumor.ttl), and defines our target diagnosis class (
:ReportOfThyroidDisease) as being anything classified in SNOMED-CT as a disorder of the thyroid gland (code sct:14304000.
-
Select
Start Reasonerunder theReasonermenu. It took ~2 minutes to run on a 3.4GHz laptop.
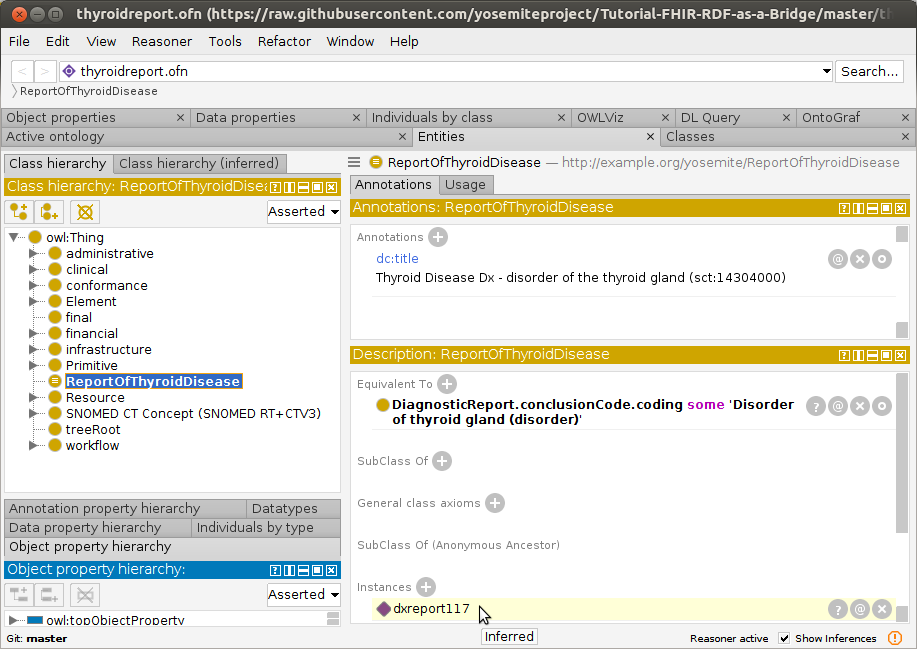
- Navigate to
ReportOfThyroidDiseasein theClass Hierarchytab and observe thatdxreport117has been classified as an instance of thyroid disease.
How it works
To further understand how this demo works, examine the roles and contents of
the files listed below.

Class definitions
These are OWL files we created to specify the kinds of diagnoses that we wish to identify, such as cancer or thyroid disease.
- fullreport.ofn – Class definition for
:FinalPatientReportWithCancerDiagnosis, which is a final patient report of cancer diagnosis. This class is the intersection of three classes defined in separate files::PatientReport:FinalReport:ReportWithCancerDiagnosis
- patientreport.ofn – Class definition for
:PatientReport, i.e., all reports whose subject is a fhir:Patient - finalreport.ofn – Class definition for
:FinalReport, i.e., all reports whose status meets our criteria for finalized - cancerreport.ofn – Class definition for
:ReportWithCancerDiagnosis, which are reports having a diagnosis of any 363346000: Malignant neoplastic disease. - thyroidreport.ofn – Class definition for
:ReportOfThyroidDisease, which are reports having a diagnosis of any 14304000: Disorder of thyroid gland (disorder). - finalreport_text.ofn – [Not used in this tutorial] Class definition for
:FinalReportwhose status text matches what we think counts as “finalized”. This is a potential alternate way of defining the:FinalReportclass.
Instance data
These files represent the FHIR medical reports that are to be analyzed to determine whether they represent the target diagnosis, such as cancer or thyroid disease. They were originally downloaded from the HL7 FHIR site, but snapshots of these files are included here to insure that this tutorial will still work correctly even if those examples are moved or modified on the HL7 site. For this reason, these files were modified to point to these github versions instead of pointing to the original versions on the HL7 site. One way to see what lines were changed is to search for the word “github” within these files.
- diagnosticreport-example-f201-brainct.ttl – This report contains a diagnosis of 188340000: Malignant tumor of craniopharyngeal duct (disorder) . Using the SNOMED-CT ontology, the reasoner will conclude that this is a kind of cancer - a 363346000: Malignant neoplastic disease.
- diagnosticreport-example-dxreport117-thyroidtumor.ttl – This report contains a diagnosis of Malignant tumor of left lobe of thyroid gland
- imagingstudy-example-xr.ttl
- imagingstudy-example-xr-mod.ttl – Imaging study with sample laterality transformation
Ontologies / vocabularies
These are standard SNOMED-CT and FHIR ontologies/vocabularies that have been downloaded for use in this analysis. Ideally these ontologies would be usable as-is after downloading them from the HL7 and IHTSDO websites. However, a few local modifications were made for this tutorial, as described below, in addition to modifying URIs to point to these github versions.
- codesystem-diagnostic-report-status.ttl – proposed OWL representation of the
DiagnosticReport.statuscode system. This mini-ontology was not downloaded from the HL7 site, because it has not yet been standardized as part of the FHIR release. However, the FHIR/RDF group is working toward including it in the FHIR release. (It needs to be integrated into the FHIR specification build process so that it is auto-generated and stays in sync with the rest of the FHIR specification.) Once it is a part of the FHIR release it will be included in the FHIR/RDF definitions. - fhir.ttl – FHIR (version R4) Metadata vocabulary included in the FHIR/RDF definitions, but with: (a) the
xsd:base64Binarydatatype changed toxsd:dateTime(to prevent the reasoner from barfing on an unknown datatype); and (b) ontology URIs changed to point to saved snapshots on github, to ensure that this tutorial will continue to work even as FHIR and SNOMED-CT evolve.- fhir_ORIGINAL.ttl – Original version of the FHIR (version R4) Metadata vocabulary as downloaded from the FHIR/RDF definitions
- fhir_diffs.txt – Differences between fhir_ORIGINAL.ttl and fhir.ttl
- w5.ttl – FHIR (version R4) 5 W’s ontology – Who, What, When, Where, Why – but with the ontology URI changed to point to a saved snapshot on github.
- w5_ORIGINAL.ttl – Original version of the FHIR (version R4) 5 W’s ontology, as downloaded from the FHIR/RDF definitions.
- w5_diffs.txt – Differences between w5_ORIGINAL.ttl and w5.ttl
- snomed_cancer_subset.ttl – an OWL representation of the transitive closure and neighborhood of concepts:
- 18834000: Malignant tumor if craniopharyngeal duct (disorder)
- 394914008: Radiology - speciality (qualifier value)
-
429858000: Computed tomography of head and neck (procedure)
See SNOMED_CT directory for description of how this subset was generated
- snomed_thyroid_subset.ttl – An OWL representation of the transitive closure of:
- 394914008: Radiology - specialty (qualifier value)
- 429858000: Computed tomography of head and neck (procedure)
- 363346000: Malignant neoplastic disease (disorder)
- 363698007: Finding site (attribute)
- 170784008: Entire left lobe of thyroid gland (body structure)
- 14304000: Disorder of thyroid gland (disorder)
See SNOMED_CT directory for description of how this subset was generated
Protege files
These files are generated by Protege. They are not needed.
- catalog-v001.xml – XML catalog used by Protege. This causes all references to be resolved locally
- catalog-v001.backup.xml – Backup copy of XML catalog as Protege tends to scribble on these things if you so much as look at it crosseyed
Acknowledgements
Thanks to Harold Solbrig for originally creating this demo, and to David Booth and Gopikrishnan (“Gopi”) Chandrasekharan for editing it into this tutorial and video.
Corrections or suggestions?
Please submit a pull request or email David Booth with “Yosemite Project – Tutorial-FHIR-RDF-as-a-Bridge” as the subject line.
